Past Members: Chenping Yu, Guilherme Ruppert, Leonid Teverovskiy, and O. Carmichael
MSP Extraction:
A New Symmetry-Based Method For Mid-Sagittal Plane Extraction In Neuroimages
Citation
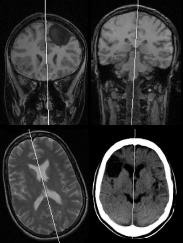 | Guilherme Ruppert and Leonid Teverovskiy and Chenping Yu and Alexandre Falcao and Yanxi Liu. A New Symmetry-Based Method For Mid-Sagittal Plane Extraction In Neuroimages, IEEE International Symposium on Biomedical Imaging (ISBI) 2011
Pages 285-288 [PDF] |
Abstract
The estimation of the mid-sagittal plane (MSP) is a known problem with several applications in neuroimage analysis. As advance
to the state-of-the-art, we present a considerably better approach for MSP extraction based on bilateral symmetry maximization and a more suitable error metric to compare MSP estimation methods. The proposed method was quantitatively evaluated using three other state-of-the-art approaches as baselines and a heterogeneous dataset with 164 clinical images. It outperformed the others in accuracy and precision, being well succeeded on all images. Besides, it does not present limitations with respect to the imaging protocol and initial position of the head, and it is one of the fastest methods in the literature, taking around 30 seconds on a regular workstation.
A Quantitative Comparison Study of Neuroimage Midsagittal Plans Extraction
Citation
 | Guilherme C. S. Ruppert and Leonid Teverovskiy and Chenping Yu and Alexandre X. Falcao and Yanxi Liu. A Quantitative Comparison Study of Neuroimage Midsagittal Plans Extraction, PSU CSE #09-014 2009. [PDF] |
Abstract
to be added
Truly 3D Midsagittal Plane Extraction for Robust Neuroimage Registration
Citation
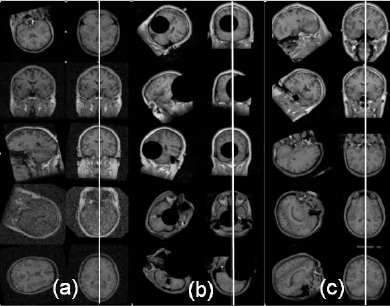 | L. Teverovskiy and Yanxi Liu. Truly 3D Midsagittal Plane Extraction for Robust Neuroimage Registration, International Symposium on Biomedical Imaging: Macro to Nano
2006, Pages 860-863
[PDF] |
Abstract
This paper describes a robust algorithm for reliable ideal Midsagittal Plane extraction (iMSP) from 3D neuroimages. The algorithm makes no assumptions about initial orientation of a given 3D brain image and works reliably on neuroimages of normal brains as well as brains with significant pathologies. Presented technique is truly three-dimensional since we treat each neuroimage as a three-dimensional volume rather than a set of two-dimensional slices. We use an edgebased approach which employs cross-correlation to extract iMSP. Proposed algorithm was quantitatively evaluated on a variety of real and artificial neuroimages. We find that our algorithm is able to extract iMSP from neuroimages with arbitrary initial orientations, large asymmetries, and low signal to noise ratio. We also demonstrate that presented algorithm can increase robustness of existing neuroimage registration algorithms, be it rigid, affine or less restricted deformable registration. Our algorithm was implemented using Insight Toolkit(ITK).
Robust Midsagittal Plane Extraction from Normal and Pathological 3D Neuroradiology images
Citation
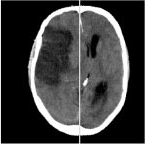 | Yanxi Liu and Robert T. Collins and W.E. Rothfus. Robust Midsagittal Plane Extraction from Normal and Pathological 3D Neuroradiology images, IEEE Transactions on Medical Imaging 2001Volume 20, Number 3, Pages 175 - 192
[PDF] |
Abstract
This paper focuses on extracting the ideal midsagittal plane (iMSP) from three-dimensional (3-D) normal and pathological neuroimages. The main challenges in this work are the structural asymmetry that may exist in pathological brains, and the anisotropic, unevenly sampled image data that is common in clinical practice. We present an edge-based, cross-correlation approach that decomposes the plane fitting problem into discovery
of two-dimensional symmetry axes on each slice, followed by a robust estimation of plane parameters. The algorithm’s tolerance to brain asymmetries, input image offsets and image noise is quantitatively evaluated. We find that the algorithm can extract the iMSP from input 3-D images with 1) large asymmetrical lesions; 2) arbitrary initial rotation offsets; 3) low signal-to-noise ratio or high bias field. The iMSP algorithm is compared with an approach based on maximization of mutual information registration, and is found to exhibit superior performance under adverse conditions. Finally, no statistically significant difference is found between the midsagittal plane computed by the iMSP algorithm and that estimated by two trained neuroradiologists.
Classification-Driven Pathological Neuroimage Retrieval Using Statistical Asymmetry Measures
Citation
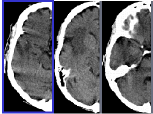 | Yanxi Liu and F. Dellaert and W.E. Rothfus and A. Moore and J. Schneider and T. Kanade. Classification-Driven Pathological Neuroimage Retrieval Using Statistical Asymmetry Measures,
Medical Imaging Computing and Computer Assisted Intervention (MICCAI) 2001.
[PDF] |
Abstract
This paper reports our methodology and initial results on volumetric pathological neuroimage retrieval. A set of novel image features are computed to quantify the statistical distributions of approximate bilateral asymmetry of normal and pathological human brains. We apply memory-based learning method to find the most-discriminative feature subset through image classification according to predefined semantic categories. Finally, this selected feature subset is used as indexing features to retrieve medically similar images under a semantic-based image retrieval framework. Quantitative evaluations are provided.
Evaluation of a Robust Midsagittal Plane Extraction Algorithm for Coarse, Pathological 3D Images
Citation
 | Yanxi Liu and Robert T. Collins and W.E. Rothfus. Evaluation of a Robust Midsagittal Plane Extraction Algorithm for Coarse, Pathological 3D Images,
Medical Imaging Computing and Computer Assisted Intervention (MICCAI) 2000.
[PDF] |
Abstract
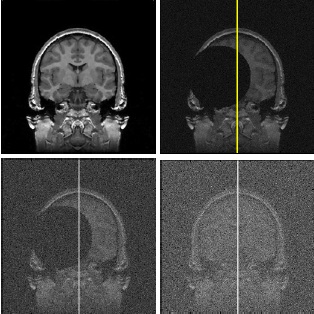
This paper focuses on the evaluation of an ideal midsagittal plane (iMSP) extraction algorithm. The algorithm was developed for capturing the iMSP from 3D normal and pathological neural images. The main challenges are the drastic structural asymmetry that often exists in pathological brains, and the sparse, nonisotropic data sampling that is common in clinical practice. A simple edge-based, cross-correlation approach is presented that decomposes the iMSP extraction problem into discovery of symmetry axes from 2D slices, followed by robust estimation of 3D plane parameters. The algorithm's tolerance to brain asymmetries, input image offsets and image noise is quantitatively measured. It is found that the algorithm can extract the iMSP from input 3D images with (1) large asymmetrical lesions; (2) arbitrary initial yaw and roll angle errors; and (3) low signal-to-noise level. Also, no significant difference is found between the iMSP computed by the algorithm and the midsagittal plane estimated by two trained neuroradiologists.
Automatic Bilateral Symmetry (Midsagittal) Plane Extraction from Pathological 3D Neuroradiological Images
Citation
 | Yanxi Liu and Robert T. Collins and W.E. Rothfus. Automatic Bilateral Symmetry (Midsagittal) Plane Extraction from Pathological 3D Neuroradiological Images,
SPIE International Symposium on Medical Imaging 1998.
[PDF] |
Abstract
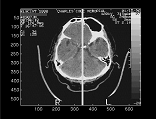
Most pathologies (tumor, bleed, stroke) of the human brain can be determined by a symmetry-based analysis of neural scans showing the brain's 3D internal structure. Detecting departures of this internal structure from its normal bilateral symmetry can guide the classification of abnormalities. This process is facilitated by first locating the ideal symmetry plane (midsagittal) with respect to which the brain is invariant under reflection. An algorithm to automatically identify this bilateral symmetry plane from a given 3D clinical image has been developed. The method has been tested on both normal and pathological brain scans, multimodal data (CT and MR), and on coarsely sliced samples with elongated voxel sizes.
Alzheimer's Disease Diagnosis
Quantified Brain Asymmetry for Age Estimation of Normal and AD/MCI Subjects
Citation
 | Leonid Teverovskiy and James Becker and Oscar Lopez and Yanxi Liu. Quantified Brain Asymmetry for Age Estimation of Normal and AD/MCI Subjects,
SPIE International Symposium on Medical Imaging 2008.
[PDF] |
Abstract
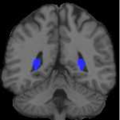
We propose a quantified asymmetry based method for age estimation. Our method uses machine learning to discover automatically the most discriminative asymmetry feature set from different brain regions and image scales. Applying this regression model on a T1 MR brain image set of 246 healthy individuals (121 females; 125 males, 66 ± 7.5 years old), we achieve a mean absolute error of 5.4 years and a mean signed error of -0.2 years for age estimation on unseen MR images using the stringent leave-15%-out cross validation. Our results show significant changes in asymmetry with aging in the following regions: the posterior horns of the lateral ventricles, the amygdala, the ventral putamen with a nearby region of the anterior inferior caudate nucleus, the basal forebrain, hyppocampus and parahyppocampal regions. We confirm the validity of the age estimation model using permutation test on 30 replicas of the original dataset with randomly permuted ages (with p-value < 0.001). Furthermore, we apply this model to a separate set of MR images containing normal, Alzheimer’s disease (AD) and mild cognitive impairment (MCI) subjects. Our results reflect the relative severity of brain pathology between the three subject groups: mean signed age estimation error is 0.6 years for normal controls, 2.2 years for MCI patients, and 4.7 years for AD patients.
Discovery of "Biomarkers" for Alzheimer's Disease Prediction from Structural MR Images
Citation
 | Yanxi Liu and L. Teverovskiy and O. Lopez and H. Aizenstein and C. Meltzer and J. Becker. Discovery of "Biomarkers" for Alzheimer's Disease Prediction from Structural MR Images,
SPIE International Symposium on Medical Imaging 2007.
[PDF] |
Abstract
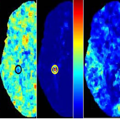
We propose a computational framework for learning predictive image features as “biomarkers” for Alzheimer’s Disease discrimination using high-resolutionMagnetic Resonance (MR) brain images. We focus on the exploration of a very large (> 500 million) feature space derived extensively from the deformation and tensor ??elds. In such a huge space, our computational tool supports an automatic search for discriminative feature subspaces and the corresponding anatomical regions in human brains, which can be used to discriminate previously unseen, individual structuralMR images from Alzheimer’s Disease (AD) and normal control (CTL) subjects. Our aggressive leave-ten-out cross-validations on 40 subjects demonstrate higher than 90% sensitivity and specificity. In addition, we demonstrate intriguing anatomical locations as automatically discovered “biomarkers” and the spatial distributions of 20 Mild cognitive impairment (MCI) subjects in the discriminative feature space automatically learned for AD and CTL separations. Our results illustrate a truly complementary effort of human and computers for early diagnosis of AD from MR images.
Atlas-Based Hippocampus Segmentation in Alzheimer's Disease and Mild Cognitive Impairment
Citation
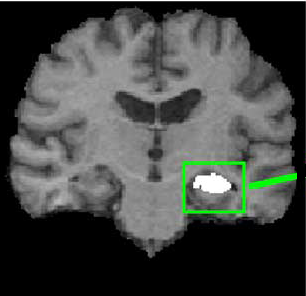 | O. Carmichael and H. Aizenstein and S.W. Davis and J. Becker and P.M. Thompson and C. Meltzer and Yanxi Liu. Atlas-Based Hippocampus Segmentation in Alzheimer's Disease and Mild Cognitive Impairment,
NeuroImage 2005, Number 27, Pages 979 - 990.
[PDF] |
Abstract
This study assesses the performance of public-domain automated methodologies for MRI-based segmentation of the hippocampus in elderly subjects with Alzheimer’s disease (AD) and mild cognitive impairment (MCI). Structural MR images of 54 age- and gendermatched healthy elderly individuals, subjects with probable AD, and subjects with MCI were collected at the University of Pittsburgh Alzheimer’s Disease Research Center. Hippocampi in subject images were automatically segmented by using AIR, SPM, FLIRT, and the fully deformable method of Chen to align the images to the Harvard atlas, MNI atlas, and randomly selected, manually labeled subject images (‘‘cohort atlases’’). Mixed-effects statistical models analyzed the effects of side of the brain, disease state, registration method, choice of atlas, and manual tracing protocol on the spatial overlap between automated segmentations and expert manual segmentations. Registration methods that produced higher degrees of geometric deformation produced automated segmentations with higher agreement with manual segmentations. Side of the brain, presence of AD, choice of reference image, and manual tracing protocol were also significant factors contributing to automated segmentation performance. Fully automated techniques can be competitive with human raters on this difficult segmentation task, but a rigorous statistical analysis shows that a variety of methodological factors must be carefully considered to insure that automated methods perform well in practice. The use of fully deformable registration methods, cohort atlases, and user-defined manual tracings are recommended for highest performance in fully automated hippocampus segmentation.
Atlas-Based Hippocampus Segmentation in Alzheimer's Disease and Mild Cognitive Impairment
Citation
 | O. Carmichael and H. Aizenstein and S.W. Davis and J. Becker and P.M. Thompson and C. Meltzer and Yanxi Liu. Atlas-Based Hippocampus Segmentation in Alzheimer's Disease and Mild Cognitive Impairment,
CMU-RI-TR-04-53 2004.
[PDF] |
Abstract
Purpose. To assess the performance of standard image registration techniques for automated MRI-based segmentation of the hippocampus in elderly subjects with Alzheimer’s Disease (AD) and mild cognitive impairment (MCI). Methods. Structural MR images of 54 age- and gender-matched healthy elderly individuals, subjects with probable AD, and subjects with MCI were collected at the University of Pittsburgh Alzheimer’s Disease Research Center. Hippocampi in subject images were automatically segmented by using AIR, SPM, FLIRT, and the fully-deformable method of Chen to align the images to the Harvard atlas, MNI atlas, and randomly-selected, manually-labeled subject images. Mixed-effects statistical models analyzed the effects of side of the brain, disease state, registration method, choice of atlas, and manual tracing protocol on the agreement between automated segmentations and expert manual segmentations. Results. Registration methods that produced higher degrees of geometric deformation produced automated segmentations with higher agreement with manual segmentations. Automated-manual agreement between Chen’s method and expert manual segmentations were competitive with manual-manual agreement. Segmentations of the right hippocampus were more consistent with manual segmentations than those of the left. Automated-manual agreement was significantly lower in AD brains than MCI or controls. Automated segmentations based on registration with a randomly-selected subject image were more consistent with manual segmentations than those based on registration with the Harvard or MNI atlas. The manual tracing protocol was a significant source of variation in automated-manual agreement.
Discriminative MR Image Feature Analysis for Automatic Schizophrenia and Alzheimer's Disease Classification
Citation
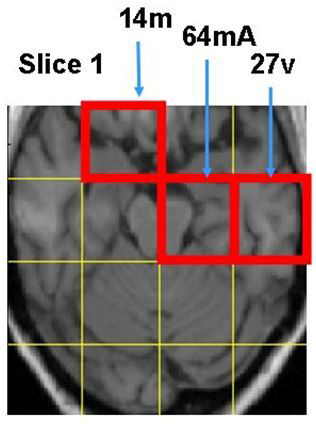 | Yanxi Liu and L. Teverovskiy and O. Carmichael and R. Kikinis and M. Shenton and C.S. Carter and V.A. Stenger and S. Davis and H. Aizenstein and J. Becker and O. Lopez and C. Meltzer. Discriminative MR Image Feature Analysis for Automatic Schizophrenia and Alzheimer's Disease Classification,
Proceedings of the 7th International Conference on MedicalImage Computing and Computer Aided Intervention (MICCAI) 2004. Pages 393 - 401
[PDF] |
Abstract
We construct a computational framework for automatic central nervous system (CNS) disease discrimination using high resolution Magnetic Resonance Images (MRI) of human brains. More than 3000 MR image features are extracted, forming a high dimensional coarseto-fine hierarchical image description that quantifies brain asymmetry, texture and statistical properties in corresponding local regions of the brain. Discriminative image feature subspaces are computed, evaluated and selected automatically. Our initial experimental results show 100% and 90% separability between chronicle schizophrenia (SZ) and first episode SZ versus their respective matched controls. Under the same computational framework, we also find higher than 95% separability among Alzheimer’s Disease, mild cognitive impairment patients, and their matched controls. An average of 88% classification success rate is achieved using leave-one-out cross validation on five different well-chosen patient-control image sets of sizes from 15 to 27 subjects per disease class.
links to earlier work













